Background
Protein A chromatography is commonly used for the affinity capture of antibodies directly from harvested cell culture fluid (HCCF). The highly specific interaction between the protein A ligand and the Fc portion of antibodies and Fc fusion molecules enables significant purification factors to be achieved. The mechanism for clearance of viruses on protein A chromatography is that viruses largely flow through the column and are removed predominately in the non-binding flow-through fraction during the loading and wash out phases of the chromatography operation. Although the vast majority of virus is non-binding, a small fraction of virus is retained by the protein A resin matrix and co-elutes with product during the pH transition that is used to effect elution. The nature of the non-specific interaction of the trace amounts of virus that bind to the resin matrix is not well understood. As a consequence, the log reduction values (LRVs) that are obtained for this step are often variable from product to product, ranging typically from two to greater than four log10 of clearance (1, 2). However, it is also noted that for a given product and process, the LRV values obtained are actually highly reproducible.
A second mechanism of clearance on protein A chromatography is the inactivation of susceptible virus (generally enveloped virus) during the low-pH conditions used to elute the product. The extent of low-pH inactivation is related to the both the pH and exposure time. Assuming that the pH and exposure time is well controlled during production, one can claim both physical removal by protein A and inactivation by low pH on this step as long as no other low-pH inactivation unit operations are claimed in the production process. Most commonly, antibody and Fc fusion processes use a separate and dedicated low-pH inactivation unit operation to effect the inactivation of enveloped virus. In these cases, the inactivation by low pH across the protein A step should not be claimed. Utilizing different assays (i.e., infectivity and quantitative polymerase chain reaction [Q-PCR]) to deconvolute the contributions of chromatography partitioning from low-pH inactivation mechanisms are useful in this regard.
Impact of Process Parameters on Viral Clearance Results Achieved on Protein A–Based Affinity Step (Olga Galperina, David Kahn; Human Genome Sciences)
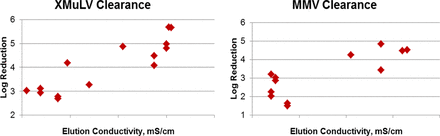
Protein A-based affinity capture can be very effective in removal of retro- and parvoviruses. However, the performance of this step is highly variable from project to project, and results as low as 2 log10 and as high as 6 log10 of clearance have been observed. There is no clear understanding of the interactions between monoclonal antibody (mAb) captured on the column, spiked viruses, and the resin matrix, which results in a lack of understanding of how to make affinity steps more robust. In order to better understand the impact of process parameters on the performance of affinity columns, results from multiple platform processes were compared. There were no correlations between achieved clearance results and parameters such as wash conductivity, elution pH, load pH, load conductivity, concentration, wash volume, and wash conductivity. A weak correlation with clearance results and wash pH and a stronger correlation with elution conductivity were observed. Based on in-house data, conductivity of elution buffer could have significant impact on clearance results for murine leukemia virus (MuLV) and mice minute virus (MMV) models (Figure 1). Higher elution conductivity can be very beneficial, as clearance results above 4 log10 were achieved. The mechanism behind this correlation, however, is still unclear.
Study by Human Genome Sciences demonstrating that conductivity of elution buffer can have significant impact on protein A virus clearance.
Key Process Parameters for Removal of Model Retrovirus and Parvovirus by Agarose-Based Recombinant Protein A Affinity Chromatography (John Mattila; Regeneron Pharmaceuticals Inc.)
An evaluation of in-house data was performed to investigate potential worst-case parameters for removal of viruses by a highly cross-linked agarose recombinant protein A resin. The investigation included the model retrovirus MuLV and the model parvovirus MMV. The data set included one resin type, and all chromatography methods had used identical buffers and step lengths.
The review supports published findings for removal of Chinese hamster ovary (CHO) endogenous retrovirus-like particles by a controlled pore glass recombinant protein A media (3). Process parameters including load volume and flow rate had a statistically significant impact on retrovirus clearance. There is a difference of approximately 1 log10 between mean retrovirus removal and removal during worst-case conditions of low load volume and low linear velocity.
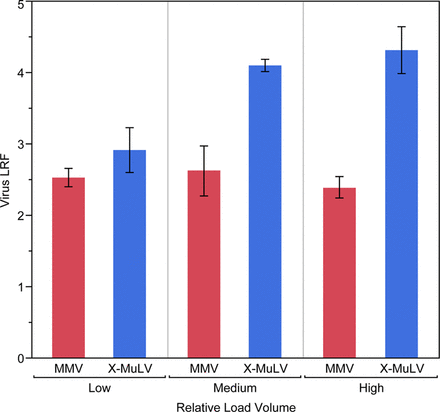
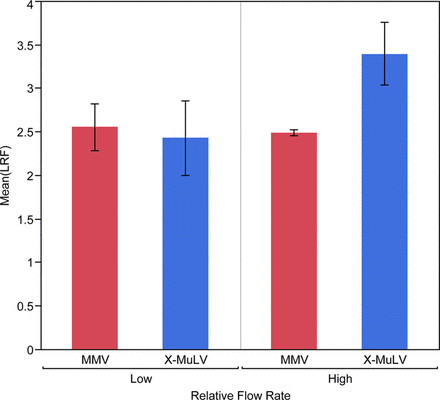
Results showed that MMV removal is consistently lower than MuLV removal and suggested that clearance is less affected by process conditions (Figures 2 and 3). Because MMV and MuLV have been compared head-to-head in each spiking study, this is the clearest result of this retrospective data review.
Retrospective review of Regeneron mAb platform performance (N = 18) reveals low volumetric column loading during recombinant Protein A (rProteinA) affinity chromatography results in a statistically meaningful reduction (P = 0.01) of xenotropic murine leukemia virus (X-MuLV) clearance.
Retrospective review of Regeneron mAb platform performance at low load volume (N= 14) reveals low flow rate during rProteinA affinity chromatography may result in a meaningful reduction (p = 0.14) of xenotropic murine leukemia virus (X-MuLV) clearance. There is no evidence that MMV clearance is reduced.
Impact of Product on MuLV Removal by Protein A Chromatography (L. Connell-Crowley; Amgen)
The behavior of model retroviruses, such as MuLV, during protein A chromatography is not well understood. In Amgen's experience, protein A can remove up to 4 log10 of MuLV but sometimes only removes as little as 1 log10 despite the fact that virus is not expected to bind to this affinity resin. Additionally, LRVs obtained through the mechanism of removal by protein A are reproducible for a given mAb, but can vary widely for different mAbs or different processing conditions. Experiments were performed to study the behavior of MuLV on a mAbSure protein A column and on the Sepharose backbone alone, in the presence or absence of mAb products and/or impurities.
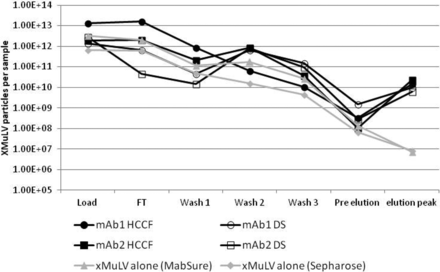
The results showed that virus alone has little to no interaction with mAbSure or the Sepharose resin backbone, as ∼5 log10 or more can be achieved. In the presence of HCCF or purified drug substance, a portion of the virus appeared to interact, and co-elute, with the mAb product, resulting in LRVs of ∼2-3 log10 (Figure 4). The apparent interaction of virus with product and/or impurities in the feedstock may explain why there is variability in LRVs across different products.
xMuLV clearance is reduced by the presence of mAb product and/or impurities in an Amgen study. Various load materials were spiked with 0.5% xMuLV and loaded onto MabSure or Sepharose columns. Fractions were collected and xMuLV quantitated by QPCR.
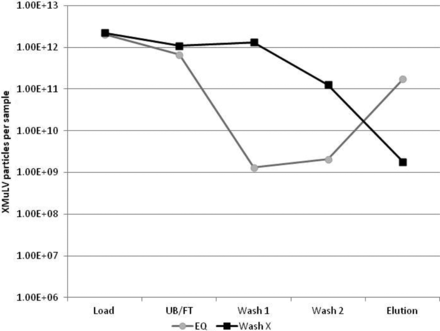
There also appears to be some room for improving MuLV removal with specific washes that can disrupt this interaction. An experiment comparing the MuLV content of in-process samples for a protein A run that was washed only with equilibration (EQ) buffer, to a protein A run that included a wash known to remove host cell protein (HCP) (Wash X), showed there was a higher MuLV content in the wash samples, and a lower MuLV content in the elution sample (Figure 5). This suggests that some of the MuLV that was interacting with the resin, the mAb product, or the impurities that had bound to the column were removed with Wash X.
QPCR data of protein A runs with 0.5% xMuLV spike in a model Amgen HCCF load, using either EQ wash only or Wash X, followed by EQ wash. Use of Wash X increases particle count in wash fractions and reduces particle count in the elution fraction, suggesting that it is removing virus during the wash steps.
- © PDA, Inc. 2014