Abstract
Microbial challenge testing is a common procedure to determine the retention efficiency, performance, and validity of a sterilizing-grade filter. The ASTM 838-05 standard describes a bacteria challenge test procedure based on Brevundimonas diminuta (ATCC 19146), routinely used to verify a 0.2 μm rated sterilizing-grade filter. Process validation procedures most often also utilize B. diminuta (ATCC 19146), but instead of the standard procedures and fluids, process, and product parameters are employed to determine whether these parameters influence the retentivity of the filter or changes to the challenge organism, which might result in the penetration of the filter. In certain instances, the native bioburden within the drug manufacturing process is used to perform such process validation challenge tests. Filter penetrations can happen and cause concern; therefore, it is essential to identify the organism species with accuracy to avoid unnecessary confusion. This paper and its follow-up will describe such imprecision and the resulting misconceptions. It will clarify past determinations and put perspective on the findings.
LAY ABSTRACT: Sterilizing-grade filters are used to remove microorganisms from biopharmaceutical solutions. To determine the retention performance of such filters, bacteria challenge tests are utilized, often with a standard challenge organism (Brevundimonas diminuta), in instances with native bioburden. The accuracy of the microorganism identification is of importance to avoid flawed results and misinterpretation of the filter's performance.
- Microbial challenge test
- Sterilizing grade filter
- Hydrogenophaga pseudoflava
- Curvibacter
- Process validation
Introduction
Sterilizing-grade membrane filters, commonly rated as 0.2 μm filters, are of essential importance within aseptic processes to remove contaminants, mainly microorganisms. Because these filters may represent the last microbial barrier before filling of the drug product, the retentive performance of such filters needs to be proven. Product and process validation of the filter utilizing the process parameters and, if possible, the drug product is expected by regulatory authorities. Viability tests are part of the process validation to assure the challenge organism stays alive within the challenge fluid under the process conditions. The viability test is followed by a bacteria challenge test, if possible with the actual drug product, and if not, with the closest possible surrogate utilizing the process conditions. The most commonly used microorganism for such challenge tests is Brevundimonas diminuta (ATCC 19146), established within ASTM 838-05 (1) and furthermore mentioned in guidance documents like the Food and Drug Administration (FDA) Aseptic Processing Guidance for Industry (2) and PDA Technical Report No. 26 (3). The process validation activities are required to evaluate whether or not the challenge organism can penetrate the sterilizing-grade filter chosen. Organisms can penetrate 0.2 μm rated filters due to effects of the fluid or process conditions on the filter or microorganism. It has to be established that there are no effects such as organism shrinkage or filter membrane damage due to fluid or process parameters.
In 1999, rare microorganism penetration became of increased importance and calls were made to switch the pore size rating of sterilizing-grade filters from 0.2 to 0.1 μm (4). These suggestions were enforced by challenge tests of 0.2 μm rated filters with a supposedly newly discovered challenge organism identified by FAME (fatty acid methyl ester) analysis as Hydrogenophaga pseudoflava (ATCC 700892) (5–8). Fortunately, appropriate scientific discussions following these implicative publications focused on the need to utilize the best approach to process- and product-related validation instead of sole reliance on pore size ratings. In light of this, recent publications showed challenge tests performed to support Sundaram's papers (9, 10). The same organism, specified as H. pseudoflava (ATCC 700892), was used within these studies. The authors claim that H. pseudoflava challenge tests might be a good challenge organism choice to test filtration parameter impact on 0.2 μm rated filters. Unfortunately, the studies were performed with laboratory or analytical filter membranes and not filters used in production processes, which render the data interesting but of no practical use because only pleated filter devices are used in production settings. Moreover, all mentioned publications specifying this organism utilized it incorrectly for challenge testing. Throughout all mentioned publications, the challenge organism ATCC 700892 was named as H. pseudoflava but in reality has been phylogenetically affiliated to the genus Curvibacter, as shown in this paper.
As the data from Sundaram et al. (5) show, the organism ATCC 700892 is adaptable and is of smaller size, especially in diameter. The organism has a curved, thin shape, and it aligned properly to the pore structure to penetrate filter membranes. According to Sundaram et al., the strain ATCC 700892 phylogenetically incorrectly classified as H. pseudoflava has been suspected as a problem organism, penetrating 0.2 μm rated membrane filters, so-called sterilizing-grade filters. The species H. pseudoflava may be more frequently found in biopharmaceutical production settings than Curvibacter. In contrast, the genus Curvibacter, truly the strain ATCC 700892, has to our knowledge rarely been found within biopharmaceutical production settings. Challenge tests with the type strain H. pseudoflava (ATCC 33668) show that the organism will be retained by 0.2 μm rated membrane filters (results in the forthcoming Part II of this series of articles).
This shows that the exact determination of the challenge organism is of importance, moreover, when the significance of the challenge organism within production settings is an essential part of typical risk analysis. The question of the existence of diminutive organisms has never been in question. The question has to be, does one find such organisms within the process stream and what impact would such a finding have? It is not of value to challenge filters randomly with any organism found, but to proceed with appropriate product- and process-related validation work to show that the specific filter will perform as desired within a specific process and product stream.
Methods
For the phylogenetic characterization, strain ATCC 700892 was analysed by sequencing of the 16S rRNA gene. The nearly full-length 16S rRNA gene (1424 bp) was sequenced twice by LGC Genomics GmbH Berlin. The sequence of the strain ATCC 700892 was submitted to the NCBI GenBank (11).
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of the strain ATCC 700892 is HM357758. Phylogenetic affiliation was analysed with the ARB (latin, “arbor” = tree) software (12) using the SILVA rRNA Databases, release 102 (13), and data were compared to the NCBI GenBank using the BLAST nucleotide database.
The phylogenetic tree was calculated with partial 16S rRNA gene sequences (1407 bp; E. coli position 54-1461) by using the neighbor joining (Jukes-Cantor and Felsenstein correction), maximum parsimony, and maximum likelihood methods implemented in ARB. Different filters were used for the calculations.
The phylogenetic distances were determined by calculation of a similarity matrix with neighbor joining implemented in ARB.
The Model Challenge Organism for 0.2 μm Rated Filters
B. diminuta (ATCC 19146) has been designated as the model for those organisms expected to be found in pharmaceutical processing contexts. Studies involving organisms are often designed around the use of a model microbe. The diversity of the numerous types and sizes of microorganisms is simply too large to practically investigate each and every type that may be encountered in pharmaceutical settings. Given the large number of organism types, it is advantageous to select a model to serve for all the others, or at least for those commonly detected within pharmaceutical operations. In the process validation effort, attempts have been made to utilize representatives of the native bioburden for this purpose, having been described as a valuable risk assessment activity (3, 14). The native microbes may be difficult to culture; their life cycles may be too brief for testing purposes and they may lack ease and safety in handling, etc. In the actual event, B. diminuta has served adequately, although not universally, as a model for organism retentions by filters. The success of selecting other microbes to better fill the model role in place of B. diminuta is of low probability. The type strain H. pseudoflava (ATCC 33668) could take on such role, but why change when the experience base with B. diminuta is multiple factors higher? Change for the sake of change has not yet been seen to be of value.
The key factor governing particle removal by filters is that it involves the sieve retention mechanism wherein the size and shape of the particle, whether organism or otherwise, depends upon it being too large to pass through the filter's pores. This retention mechanism does not necessarily match all organisms potentially present in pharmaceutical processes or utilized as a challenge organism. For example, if the molecular structure of the organism's surface should incline it to interact by adsorption to the filter's polymeric surface, as by hydrogen bonding, factors other than sieve retention could intrude on the results. The modular action ascribed to the organism's size and shape in relation to the size and shape of the filter's pores could be misinterpreted. Just such a situation accounts for Ridgway's (15) observation that Pseudomonas aeruginosa organisms are more strongly retained by polyamide membranes than by cellulose triacetate filters. It also explains the removal of latex particles from aqueous suspensions by polyamide membranes in the presence of surfactant, but not in its absence (Table I) (16). Adsorption propensities, not size and shape effects on sieving, may account for these occurrences. For this reason, it is of importance to know exactly what organism is utilized for challenge tests. All organisms have different structural as well as physicochemical properties that will behave in accordance to the membrane properties.
Retention (%) of 0.198 μm Spheres by Various 0.2 μm Rated Membranes
That the adsorptive sequestration mechanism can operate to remove organisms and other particulates from fluid suspensions is well understood (17). However, the common and dominant view of particle removal by filters is that sieve retention, also called size exclusion, alone suffices to explain the retention phenomenon. Consequently, the choice of the model organism is strongly based on the microbe's size relative to the filter's pore size and shape. The other particle capture mechanisms are, thus, passed over as being subsidiary, and are afforded a lesser significance than the basis of size. The relegation of another important retention attribute to a subsidiary role encourages the bacterial challenge focus to be more on the size of the organism. This relegation of another retention mechanism creates a diminution of retention influences due to organism and membrane surface chemistries (18). The FDA's proposal of 1995 (19) asks for organism challenge tests of sterilizing-grade filters with either the actual product or a placebo under process conditions. One reason for this challenge requirement is to find out whether or not adsorptive organism capture is influenced by either the production fluid or process parameters. The product- and production-related microorganism challenge tests not only sieve retentivity, but also adsorptive, surface chemistry, and capture. Within these validation studies, any challenge organism's retention can be measured quantitatively; it just has to penetrate the filter, though this is an undesirable event.
Challenge Organism Choice and Determination
As mentioned, challenge organism choice and determination of the right specimen is of utmost importance to avoid misinterpretations of retention performance results. The decision is most commonly determined by the process stream's bioburden, but is also based on the organism's characteristics. In certain instances the process fluid is so hostile that organisms, except bacterial spores, will not survive. The retention of bacterial spores by 0.2 μm membrane filters is a given due to the large size of the spores. If the fluid's properties are less aggressive to the organism, it could be that the fluid is subtly affecting the size and surface properties of the organism without killing it. Practical examples of microbial penetration through 0.2 μm rated filters showed that the fluid was of low nutrition rate (ultrapure water) or higher osmolarity (salt or sugar solutions), causing the organism to adapt, thus commonly shrinking in size. Lengthy filtration times elevate this effect. However, not all organisms will shrink, as their dimension is already so much reduced that any further reduction would cause mortality. Despite possible bioburden challenge tests, challenge test utilizing B. diminuta under the process conditions are recommended. B. diminuta is still the model organism to define sterilizing-grade 0.2 μm rated filters. The ultimate goal of a product- and process-related challenge test is to confirm that the filter performs as intended according to defined specifications and to establish it as a sterile filtrate. Within these test settings it is not of importance to reach any quantitative result. The result has to be the absence of any challenge organism in the filtrate, when the filter is challenged at a bioburden level of 107 per square centimeter, the ASTM standard.
Scientific attempts to quantify any challenge with B. diminuta have been done utilizing 0.45 μm rated filters (20). Likewise, such challenge tests can be performed with a multitude of organism types and filter pore size ratings, though the adsorptive properties, organism adaptation, and filter device choice requires careful consideration. Common challenge organisms in comparison to pore size rating can be seen in Table II.
Common Challenge Organisms for Specific Membrane Filter Pore Size Ratings
Most common challenge test data for process-related filter devices are required to reflect process-related obstacles. If filter units or membranes are chosen randomly, tireless and valuable work might be diminished by their irrelevance to real processes. Moreover, the organisms used for any challenge test require appropriate determination. Especially when fluid property influences on the organisms are desired assessments, one may want to use organism species that are adaptable and are not already in their smallest dimensions or starved form. The only way a fluid influencing organism size will affect a starved organism is by killing it. A shift in size increasing membrane penetration probably will not be experienced. The resulting format is predetermined, either capturing of the organism or causing the death of the organism.
Curvibacter sp. ATCC 700892
16S rRNA gene sequencing of the strain ATCC 700892 indicated that this bacterium was incorrectly named H. pseudoflava. Analysis by ARB software and comparisons with 16S rRNA gene sequences available in the NCBI GenBank show that the strain ATCC 700892 belongs to a different genus within the family Comamonadaceae.
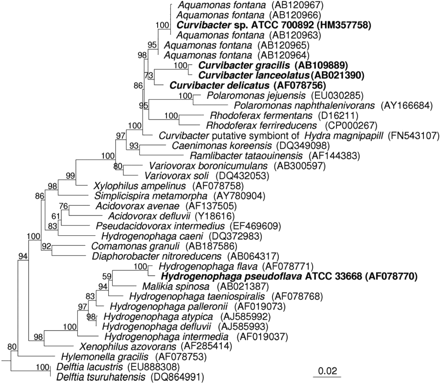
As shown in Figure 1, the strain ATCC 700892 (GenBank accession no. HM357758) has the nearest phylogenetic affiliation to Aquamonas fontana (AB120963; AB120964; AB120965; AB120966; AB120967) with a sequence similarity of 99.7 to 100.0%; however, this taxonomic name is not validly published (without valid description of the genus or species). Further phylogenetic affiliations of strain ATCC 700892 to not validly published species are not shown.
Phylogenetic neighbour joining tree (Felsenstein correction) calculated from partial 16S rRNA gene sequences (1407 bp). Bootstrap values (1000 resamplings) are indicated as percentages at branch points. Bar, 2% sequence divergence. The tree was rooted using Thermotoga maritima (GenBank accession no. M21774) as an outgroup (not shown). The strain Curvibacter sp. ATCC 700892 (HM357758) and its nearest validly published neighbours Curvibacter delicatus (AF078756), C. lanceolatus (AB021390) and C. gracilis (AB109889) as well as the type strain Hydrogenophaga pseudoflava ATCC 33668 (AF078770) are indicated in bold.
The nearest phylogenetic affiliation of the strain ATCC 700892 to a validly published species is Curvibacter delicatus (AF078756) with a sequence similarity of 98.7% as well as Curvibacter gracilis (AB109889) and Curvibacter lanceolatus (AB021390) with a sequence similarity of 98.0%. Further, a Curvibacter species as a putative symbiont of Hydra magnipapillata (FN543107) has a sequence similarity of 98.2% with the strain ATCC 700892.
Validly published species from other genera with Rhodoferax fermentas (D16211) and Variovorax boronicumulans (AB300597) have a sequence similarity of 97.0% with the strain ATCC 700892. All other validly published species show sequence similarities of <97.0%.
The sequence similarity of the strain ATCC 700892 with the type strain H. pseudoflava ATCC 33668 (AF078770) is only 94.4%. The phylogenetic distance between these two strains is clearly visible in Figure 1. The tree shows a large quantity of confirmed, published species that separate these two microorganisms. The species listed in Figure 1 represent only a selection of species due to the high data volume available, which would burden the graphic.
We propose that the strain ATCC 700892 represents a Curvibacter species and cannot be phylogenetically affiliated to H. pseudoflava. The slightly curved, rod-shaped cell morphology of the strain ATCC 700892, microscopically observed in our laboratory (data not shown) and described from Sundaram et al. (5), is comparable with the cell morphology described for the different Curvibacter species (21). This fact supports the phylogenetic affiliation of strain ATCC 700892 to the genus Curvibacter.
Curvibacter sp. ATCC 700892, wrongfully labeled as H. pseudoflava, has been utilized in multiple studies mainly to show that organisms persistently penetrate 0.2 μm rated filters (5–10). Since actually the type strain H. pseudoflava (ATCC 33668) has been retained by a multitude of 0.2 μm rated filters, the results will be listed in the forthcoming Part II of this paper; it is of importance to correct this error. As one sees, organism species play an important role when filter challenge tests are performed and especially when penetration events are interpreted. According to their sizes (Table III), B. diminuta as a stringent challenge organism is close to Curvibacter sp. ATCC 700892, whereby H. pseudoflava is larger, which is underlined by the retention findings within the studies performed. However, test conditions in low nutrient scenarios showed that the diameter of Curvibacter sp. ATCC 700892 is reduced to a significantly greater degree than B. diminuta (6). This might be the reason why this organism rather than B. diminuta penetrates a 0.2 μm rated filter. The curved rod shape might also be of influence. The specified standard sizes of the described Curvibacter species suggest that a shift of the cell diameter under low nutrient conditions is conceivable.
Size Chart of Curvibacter sp. ATCC 700892, Curvibacter delicatus ATCC 14667, Curvibacter lanceolatus ATCC 14669, Curvibacter gracilis ATCC BAA-807, Brevundimonas diminuta ATCC 19146, Hydrogenophaga pseudoflava ATCC 33668
Summary
Product- and process-related bacteria challenge tests are routinely performed to observe the retentivity assurance of sterilizing-grade filters. These challenge tests are run with the actual drug product, if possible, and under process conditions. Commonly the model challenge organism is B. diminuta, and although not a universal challenge organism it has been chosen as one of the most challenging in terms of retention and is comparatively easy to cultivate. Product- and process-related retention studies have one goal: to determine whether a specific filter achieves a sterile effluent under specific product and process circumstances. The result desired is qualitative.
Scientific studies have been performed to reach quantitative results: in some instances to observe any process- or fluid-related performance influences on retentivity, in others to promote 0.1-μm rated filters. The results desired are quantitative. The organism ATCC 700892 chosen within the mentioned studies has been erroneously named H. pseudoflava. Sequence analysis of the 16S rRNA gene of the strain ATCC 700892 showed that the organism is phylogenetical affiliated to the genus Curvibacter and the type strain H. pseudoflava (ATCC 33668) will be retained by 0.2 μm rated filters (data in the forthcoming Part II of this series).
Microorganism challenge tests are utilized to define the retentivity and often the type of membrane filter. Pore size ratings offer an administrative label or description of a particular filter type, but organism challenge tests determine the true retentive performance of the filter. To assure correct challenge test data, it is essential to determine the correct challenge organism species, as size, surface chemistries, and growth behaviors play a significant role in microorganism separation by filtration. In addition, there are organisms of significance which can be found frequently in biopharmaceutical process streams. Other organisms may survive in environments other than biopharmaceutical fluids, but these organisms are commonly not used for filter challenge tests. This paper shows the impact of incorrect organism specification, which could have led to a wrongful shift of sterilizing-grade pore size ratings. The incorrect specification caused unnecessary concerns and showed faulty penetration data.
Conflict of Interest Declaration
The author(s) declare that they have no competing interests.
Acknowledgments
We thank Martina Mehrhold and Anja Schurmann for their valuable technical assistance as well as all the personnel of the Quality Assurance Microbiology laboratory of Sartorius Stedim Biotech GmbH, Goettingen, Germany.
- ©PDA, Inc. 2011