Abstract
A method for rapid detection of microbial detection is presented. It uses the reduction of resazurin to resorufin as an indication of the presence of viable cells. The method is highly sensitive (limit of detection 1 CFU/mL) and rapid (detection time 180 s). A portable device that could allow the detection to be performed in the field is also described.
LAY ABSTRACT: Simple techniques to detect microbial contamination are needed. In particular, these need to be user-friendly and low-cost. In addition, field use capability is desirable. In this paper, we describe a device and method that has the above features.
Introduction
Rapid microbial detection is highly desirable for a number of applications (1). For example, biologics that are regulated by the Center for Biologics Evaluation and Research include live viruses (vaccines), living cells (blood, blood components, cell therapies), viral vectors (gene therapies), and biomolecules such as DNA (gene therapy) and proteins (vaccines and therapeutics). The application of traditional methods used to sterilize drugs or devices cannot be applied to these complex biologics, and yet, the often parenteral administration of the biologic product requires that the final product be sterile in order to prevent iatrogenic infection of the patient from microbially contaminated biologic products. Therefore, development of a rapid means to monitor sterility at various stages of the manufacturing process would both retain the purity of the product and provide real-time feedback on contamination that could save manufacturers time and money, while increasing the safety of the final product. For example, if a fermentation vessel is discovered to be contaminated prior to inoculation, it would be useful and save considerable time and labor to abort the process as soon as the contamination is detected.
Additionally, the availability of such rapid means would allow better control of the sterility even in the formulation and packaging of standard pharmaceuticals, which are often performed at the pharmacies. In this way, the appearance of tainted drugs on the market (2) could have been caught early. Furthermore, if the test is low-effort, and low-cost, it could be used even at the doctor's office to test suspicious drug batches for contamination.
Most “rapid detection” protocols refer to a time frame of ∼24 hours. For example (3), a published protocol uses polymerase chain reaction (PCR) to detect the presence of low levels of bacteria and mould in pharmaceutical samples. The method requires significant sample preparation and achieves detection in 27 h. It can detect contamination of less than 10 colony-forming units (CFU); however, it is not clear whether the detected cells were alive or dead at the time of the detection. There are other methods that are faster, for example measurements of the intrinsic fluorescence of the bacterial or yeast chromophores (4); such approaches are fast (seconds) and sensitive, with limits of detection again in the single-digit CFU range. However, it requires extensive knowledge about the environment in which the contamination is detected, so as to compensate for the high background signals.
Cell viability assays are widely used in drug discovery for the study of growth factors, cytokines, and cytotoxic agents. The fact that they can discover viable cells makes the ideal tool for rapid detection and quantification of microorganisms (1, 5). For example, methylene blue dye–based assay was used to monitor the bacterial load (5). There are several commercially available kits that can differentiate live and dead cells. These kits follow various methods as micro-plate assay (6), fluorescent-based assay (7, 8), fluorescent staining combined with Raman spectra (9) and fluorescence-activated cell sorting–based assay (10⇓–12) for the detection of physiological state of bacteria. Other assays based on intracellular adenosine triphosphate (ATP) for viable cell counting was previously reported (13). More recently, resazurin was also used for the screening of microbial extracts against Aspergillus fumigatus (14).
Resazurin is a weakly fluorescent dye and it is non-cytotoxic including bacteria, yeast and mammalian cells (15). It is used as oxidation-reduction indicator for the measurement of cell viability in given sample (16). The resazurin-based assay was used for the viability assays other than bacteria-like, human cells for clinical transplantation (17), stem cells (18), CD4 T cells (19), and malarial gametocytocidal assay (20). Also, resazurin-based assay can be used for the screening of bacteria for the radiation sensitivity (21). The resazurin-based assay gives a linear curve over a wide range of cell concentrations (22). Also, the resazurin assay is as sensitive as [3H] thymidine assay for detecting cell proliferation (23). Viable cells continuously convert resazurin to resorufin, increasing the overall fluorescence and color of the cell culture media surrounding the cells. Further, the fluorescent and colorimetric signal generated from the assay is proportional to the number of living cells in the sample.
In this work, we present a resazurin-based assay for high-sensitive, specific and rapid detection of E. coli in the given sample. The assay quantification is performed on a standard lab fluorometer in kinetic mode. Minimum sample preparation is needed. We demonstrate that our simple rapid assay resulted in measurement of 1 CFU/mL in a given sample in 180 s. This test was validated against standard plate method. Furthermore, we have developed a low-cost, portable, notebook-powered device that can be used for field measurements of viable-cell-induced fluorescent assays.
This assay is intended to simply identify if a vaible cell is present in the sample being tested. It does not identify the species or the growth phase of the cells that are detected. Consequently, its likely application will be as a rapid screen followed by more traditional plate-based tests where the culture can be grown and identified. However, having such a test in a portable, low-cost format should prove of great value to rapid screening of a variety of samples in the pharmaceutical and food industries.
Materials and Methods
Bacterial Strains, Media, Chemicals, and Reagents
E. coli NM303 cells were used for the present study. This strain was grown on Luria-Bertani (LB)-agar plate to verify the viability of the Escherichia coli NM303. Luria-Bertani broth was purchased in the form of powder from MP Biomedicals (Santa Ana, CA). The agar was purchased from Fisher Scientific. Resazurin sodium salt powder, was from Acros Organics (Fair Lawn, NJ). Poly(methylmethacrylate) cuvette (PMMA) was from BrandTech Scientific (Essex, CT). The plates were purchased from BD Biosciences (San Jose, CA). Other reagents like NaCl (Fisher Scientific), Na2HPO4, and NaH2PO4 were purchased from Sigma Aldrich (St. Louis, MO). All the reagents were autoclaved and filtered (0.2 μm) before use.
Preparation of E. coli Cells
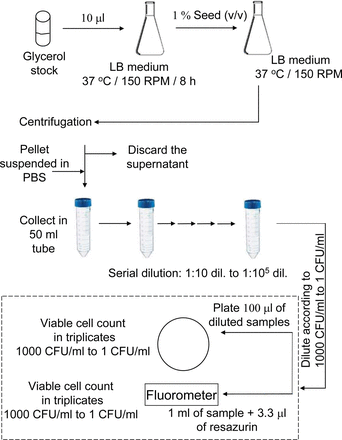
Initially, 100 mL primary culture was prepared using 10 μl of E. coli NM303 cells, which was grown at 37 °C in a shaker at 150 rpm (Lab-line Instruments, Melrose park, IL) for 8 h. The optical density of primary culture was measured to be 2.5 at 600 nm (model 8453, Agilent Technologies, Santa Clara, CA). The primary seed culture (1 %) was used to inoculate into 200 mL secondary culture were grown at 37 °C in a shaker at 150 rpm to reach an optical density of 0.4 at 600 nm (3 × 108 CFU/mL). This was centrifuged at 5000 RPM (Avanti J-25 I centrifuge, Beckman Coulter, Inc., Brea, CA) for 10 min. The cell pellet was washed with phosphate-buffered saline (PHS, pH 7.2) and the washed cells were re-suspend in PBS, pH 7.2. This sample was used for making serial dilutions (in 50 mL tubes) from 1:10 to 1:105. The 1:105 diluted sample was used to make further dilutions to make a final concentration of viable cells which was calibrated against a standard plate count. An appropriate volume was calculated (from the 1:105 aliquot) for the enumeration of 1000 CFU/mL to 1 CFU/mL and plated on LB-agar plate. It was tested for viable cell number using both standard plate count method and the resazurin reduction test. The block-schematics of the protocol are presented in Figure 1.
Experimental protocol for low CFU detection. The dashed line box represents the actual measurement.
Preparation of Resazurin Dye
For making 16 mM concentration of resazurin, 100 mg (w/w) of resazurin was weighed and suspended in 10 mL of PBS, pH 7.2. This suspension was vortexed for 30 min (5 min on/off) at room temperatute in the dark. Further, this suspension was sterile-filtered through a 0.2 μm filter, diluted to 1.6 mM, and re-vortexed for 5 min (1 min on/off). This was then filtered again and used for the present study. The sterile stock solution was stored in a cold room for a maxium of 5 days.
Plate Count Enumeration of Bacteria in a Given Sample
One milliliter aliquots (in triplicates) of sample containing cells from the serially diluted tubes were centrifuged at 13,000 rpm (accuSpin Micro 17R, Fisher Scientific), for 2 min at 4 °C. The pellet was re-suspended in 100 μL of PBS, pH 7.2 and plated on LB-agar plate. The plates were incubated at 37 °C for 24 h. CFU were counted from the each plate. A 100 μL volume of filtered PBS and heat-killed cells (negative control) was plated on LB-agar plate and incubated at 37 °C for 24 h, and no colonies were observed for the PBS and heat-killed cells.
Fluorescence Spectrocopy
The changes of the fluorescence intensity due to the reduction of resazurin were recorded using the kinetics mode of a Varian Cary Eclipse fluorescence spectrophotometer (Varian Instruments, Walnut Creek, CA). One milliliter of serially diluted cell suspension was added to a 3 mL PMMA cuvette. A 3.3 μL 1.6 mM resazurin dye was added to the cell suspension in the cuvette and mixed well. Then the cuvette was immediately placed in the fluorometer and the intensity was measured every 6 s for a period of 3 min. The excitation of the sample was performed at 530 nm and the emission was detected at 590 nm, with the excitation and emission slits set at 5 nm and the photomultiplier tube (PMT) detector voltage −750 V. The obtained readings were fitted to a straight line using the least squares method. The slopes of the fit were used as quantitaive representation of the CFU. Slopes were calculated for the intensity measured between 18 to 90 s from the start of the measurements. The assay was performed in triplicate for each standard or sample. PBS and heat-killed cell suspension was also tested as controls.
Validation of the Resazurin Reduction Test
To validate the resazurin reduction rate, several dilutions of bacteria were plated on LB-agar plates and grown for 24 h at 37 °C. The plates were analyzed for colony-forming units (CFU) with respect to each dilution. In parallel the bacterial dilutions were analyzed for resazurin reduction over a time period of 3 min. We found a good correlation between the CFU versus the rate of reduction of resazurin.
Portable Kinetics Fluorometer
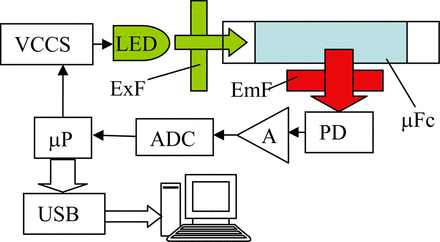
A dedicated, all-solid-state kinetics fluorometer was built in-house. In essence, this is a single-excitation, single-emission photometer that continuously measures the fluorescence intensity and plots it. The block-schematics of the fluorometer are shown in Figure 2. It utilizes semiconductor light source, an ultrabright green light-emitting diode (LED, 6 mW, SuperBrightLEDs.com) with emission maximum at 525 nm. The LED light is filtered by a bandpass interference filter 535 ± 20 nm (Intor, Soccorro, NM). The intensity of the LED is set by a voltage-controlled current source. The light is directed toward the side of a microfluidic cassette, in which the samples with the resazurin indicator are loaded. The emission is captured using a photodiode equipped with an emission filter 590 ± 20 nm. After amplification, the signal is converted in digital form and sent via universal serial bus (USB) to a computer for plotting. The data acquisition process is controlled by a microconroller.
Block-schematics of the kinetics fluorometer. VCCS—voltage-controlled current source, LED—light-emitting diode, μFc—microfluidic cassette, ExF, EmF—excitation and emission filters, PD—photodiode, A—amplifier, ADC—analog to digital converter, μP—microprocessor, USB—universal serial bus.
Microfluidics Cassestte
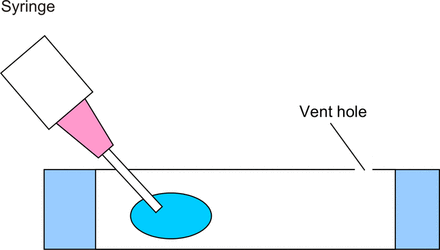
The cassette was manufactured using a laser cutter. The top and the bottom of the cassette were cut out of polycarbonate sheet and attached to the casette using pressure-sensitive adhesive. The cassette was loaded with the sample with the premixed indicator by piercing the one side of the membrane with a syringe needle, piercing the other end (see Figure 3) with the syringe and loading the sample in the cassette using the syringe.
Loading of the microfluidic cassette.
Results and Discussion
The assay described here allows for reliable detection and quatification of bacterial contamination in less than 10 min, including sample collection. We verified the results by comparing the assay with a standard plate count method. The assay was found to be fast and accurate, which makes it a good candidate for rapid detection of contamination.
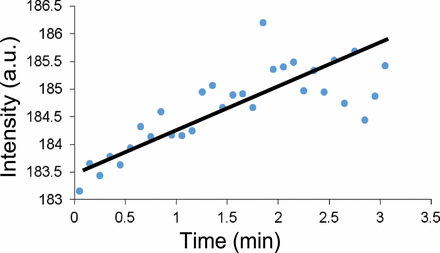
A typical kinetics curve obtained in the experiment is shown in Figure 4. The indicator (resazurin) gets reduced by the living cells, most likely by the NADH (nicotinamide adenine dinucleotide, reduced), which results in formation of a highly fluorescent compound, resorufin. By monitoring the increase of the fluorescence intensity with time, we were able to correlate the slope of the increase with the number of the viable cells. It should be noted that the magnitude of the increase is relatively small. This required working at relatively high amplifications of the fluorescence signal. Resorufin is very weakly fluorescent, and the experiment was aimed at identifying the few highly fluorescent molecules that are formed as a result of metabolic activity. The first 30 s of the monitoring were skipped—their inclusion resulted in unwanted high-magnitude oscillations of the signal which we attributed to the scattering of the light due to rapid movement of the cells (an artifact resulting from the mixing of the sample with the dye and placement of the cuvette in the fluorometer). The signal was followed for a total period of 3 min—after that the signal generally leveled off. We speculate that the reason for this is the further reduction of the resorufin to dihydroresorufin, a non-fluorescent compound.
Fluorescence intensity kinetics.
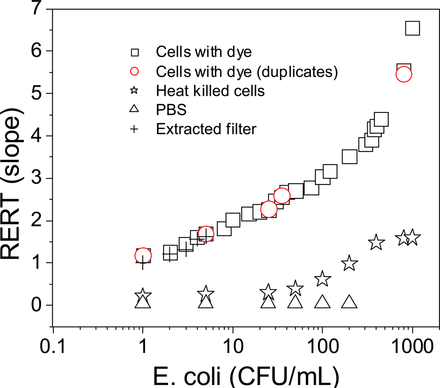
The slope of the reduction rate of resazurin was correlated to the number of bacterial cells in a given sample specifically a sample containing E. coli from 1 CFU/mL to 1000 CFU/mL and analysed in duplicate (Figure 5). There was a good correlarion between the resazurin reduction over a range of 1 CFU/mL to 1000 CFU/mL. Also, heat-killed cells were analyzed in parallel as a positive control, while the plain PBS buffer was used as a negative control. Between 1 and 100 CFU, the slope was almost zero. A slight increase in slope was observed for the heat-killed cells from 100 CFU/mL to 1000 CFU/mL. We hypothesize that this is because of the release of (fluorescent) cellular components (Figure 5). Similar results have been reported for bacteria treated with methylene blue (5), lymphocytes with alamarBlue (23), and cancer cells with Alamar Blue (24).
Correlation between the number of CFU and the slope of the fluorescence increase.
Additionally, a small number (from 1 CFU/mL to 5 CFU/mL) of cells were trapped on a filter. Then, they were reconstituted in PBS by washing the filter with PBS and analyzed. The idea behind this experiment was to see whether filtering a large volume of media that contains less than 1 CFU/mL can enrich the sample and lead to a detectable number of bacteria. The filtration enriched results showed a similar pattern of slopes compared with the directly sampled E. coli cells (Figure 5).
Further, the resazurin reduction monitoring was attempted directly in the in plain LB medium, LB medium containing cells, and LB medium containing heat-killed cells. The results indicated that the developed slopes were very high even with the plain medium. Therefore, the use of resazurin directly in the LB medium for low CFU detection should be avoided (data not shown). The LB medium by itself is a complex medium with all essential nutrients for growth. We hypothesize that this is because of enriched nutrients which can indirectly interfere with the resazurin, and this is in complete agreement with the reported data (25). We speculated that the oxidation of NADH by NADH oxidoreductase will convert the resazurin to resorufin, leading to artefacts (25).
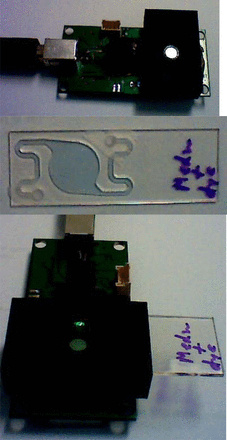
With these results, we endeavored to build a portable fluorometer coupled with a microfluidic cassette that can be used in the field for detection of contamination of media, or other contaminations (i.e., pharmaceuticals, surface contamination in the hospitals, etc.). The final design is presented in Figure 6. The electronics is powered from a USB port of a computer. This is, in essence, a single excitation, single emission fluorometer. It is designed with a cassette holder that can accept the specially designed sample cassette. The cassette is designed with relatively long input channels, which serve as a mixer. The input and the output shape of the chamber were designed to feature gradual increase and decrease of the channel width. This prevented formation of bubbles. The light enters the cassette sideways, and the wide-area photodiode picks up the fluorescence from the sample. A visualization program written in Labview provides the timing for the fluorescence acquisition. It also can calculate the slope value with every incoming reading and saves the data for further analysis on the computer's internal drive.
Portable kinetics fluorometer.
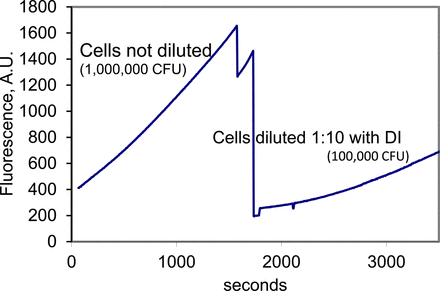
A protocol for field use of the device was developed with a goal of simplicity. Only a syringe with a syringe filter in addition to the cassette and the portable fluorometer was used. First, the sample was aspirated using a syringe equipped with 0.2 μm filter. As a result, the cells were trapped on the filter. Then, the syringe was detached from the filter, the media was discarded, and the syringe was washed with deionised water (DI) and filled with 1 mL of resazurin dissolved in DI. The syringe was reattached to the filter and the solution injected into the cassette. In the process, the cells on the filter are reconstituted in the indicator solution. The cassette was immediately paced in the portable fluorometer. The results of the device used for cell detection are presented in Figure 7. Clearly, the device works in a fashion similar to the operation of the cuvette and the fluorometer, showing different slopes with different cell concentrations. However, sensitivity of the device was lower than the sensitivity of the bench fluorometer—only cell concentrations above 10000 CFU could be detected. The reason for that is the insufficient amplification by the photodiode signal, as well as the low intensity of the light source (an LED). Future work on the device will include significant increase of the amplification as well as operating of the LED source in pulsed mode with high current amplitude (i.e., 100 times higher than the maximum DC current) and low duty ratio (i.e., 1% of the time on, 99% of the time off).
Detection of different cell concentrations using the portable fluorometer.
Conclusion
We have shown that resazurin can be used to detect the very low to high bacterial levels of bacterial contamination. The fluorescence kinetics slope over a period of time shows good correlation with the number of CFU the bacterial populations from 1 CFU/mL to 1000 CFU/mL. The low detection limit, the simplicity and rapidness of the test, as well as the fact that it is amenable for use in the field together with the availability of a simple portable device with microfluidics for its application makes it a promising approach for rapid detection of contamination.
Conflict of Interest Declaration
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by funding from FDA Grant 1U01FD004275 to the National Institute for Pharmaceutical Technology and Education. We thank Sal Nimer and Michael Duffy for making the cassettes.
- © PDA, Inc. 2014